Abstract
The formation of chimeric entities through colony fusion has been hypothesized to favour colonisation success and resilience in modular organisms. In particular, it can play an important role in promoting the invasiveness of introduced species. We studied prevalence of chimerism and performed fusion experiments in Mediterranean populations of the worldwide invasive colonial ascidian Didemnum vexillum. We analysed single zooids by whole genome amplification and genotyping-by-sequencing and obtained genotypic information for more than 2,000 loci per individual. In the prevalence study, we analysed nine colonies and identified that 44% of them were chimeric, composed of 2–3 different genotypes. In the fusion experiment 15 intra- and 30 intercolony pairs were assayed but one or both fragments regressed and died in ~45% of the pairs. Among those that survived for the length of the experiment (30 d), 100% isogeneic and 31% allogeneic pairs fused. Fusion was unlinked to global genetic relatedness since the genetic distance between fused or non-fused intercolony pairs did not differ significantly. We could not detect any locus directly involved in allorecognition, but we cannot preclude the existence of a histocompatibility mechanism. We conclude that chimerism occurs frequently in D. vexillum and may be an important factor to enhance genetic diversity and promote its successful expansion.
Similar content being viewed by others
Introduction
Natural chimerism is a widely documented phenomenon occurring in multiple phyla of protists, fungi, plants and animals, including chordates such as ascidians and mammals1. Genetic heterogeneity within organisms represents an evolutionary challenge, as several potential risks and advantages varying among taxa have been suggested but rarely tested2,3,4. Among marine invertebrates, chimerism and allorecognition have been studied in the main groups with colonial or modular species: Cnidaria5,6, Tunicata7, Porifera8,9 and Bryozoa10. These organisms show highly polymorphic histocompatibility systems which determine the output of conspecific interactions11,12. Chimeras may last for the lifetime of the colony13 or only for a few days, depending on the compatibility of the contacted colonies11. Besides the increase of genetic variability, chimerism in marine invertebrates can provide multiple benefits (e.g. enhanced growth rates, reproduction, survivorship, competition and environmental tolerances) but also significant disadvantages (e.g. developmental instability, somatic and germ cell parasitism)4,14.
Within tunicates, chimera formation in botryllid ascidians is the most studied system, particularly in Botryllus schlosseri15. In this system, a single gene locus with multiple alleles determines the outcome of the colony contacts. Colonies are usually heterozygous at this locus and they fuse when sharing at least one allele16, which means fusion between colonies occurs when they are genetically similar. Following fusion, the somatic and germ-line components of the composite unit may compete with variable outcomes17,18,19,20. In B. schlosseri there is no evidence of an improvement in growth rates, reproduction or survivorship associated to chimerism, so other ecological or evolutionary advantages should favour chimerism in this species21. Botryllid ascidians possess a common vascular system that mediates the fusion/rejection outcomes of intercolony contacts. Other colonial ascidians (e.g., didemnids), however, lack colonial blood vessels. Without vascular connections, the scope for exchange of stem cells and cell lineage competition is greatly reduced, which seemingly reduces the potential for strict colony specificity and favours more indiscriminate fusion between colonies22,23.
In invasive populations where the low genetic diversity caused by the founder effect is initially a disadvantage24, chimerism may be boosted. Increased fusion rates among different colonies could result in higher genetic diversity and richer gene expression patterns, promoting the invasiveness of the species and turning the disadvantage into an advantage for the founder population13. Moreover, each genotype from a chimeric colonial individual may adapt better to different conditions in changing environments, enhancing colony survival9,19.
The colonial ascidian Didemnum vexillum is a worldwide invasive species that has colonized most temperate regions (see25, and references therein). It can form large colonies on either natural or artificial substrates, and it can overgrow other invertebrate species such as commercial bivalves in aquaculture facilities, causing important ecological and economic loses26,27,28. D. vexillum can form chimeric colonies29 and it can also reproduce asexually by natural or human-mediated fragmentation, which is probably a major enhancer of its spread26. Its reattachment capability and fragment viability contribute to its invasive success29,30,31. Chimerism has also been suggested as a driving mechanism of the species’ remarkable invasiveness32,33.
The study of chimerism in ascidians has relied on different techniques. Monitoring of colonies in the field allows the assessment of natural fusion rates34,35,36. Chimeras can also be induced experimentally by putting in contact colonies, either growing edges or cut surfaces15,37 and examining the outcomes in the laboratory. Field and laboratory studies should ideally be complemented with genetic analyses to demonstrate intracolony heterogeneity or to characterize interacting partners. Chimeric colonies can be detected by analysing different fragments of the same colony using several genetic techniques. One of the most used methods is microsatellite genotyping, applied to ascidians38,39,40,41,42, cnidarians43 and sponges9. This kind of studies may underestimate the prevalence of chimerism, as it can only be detected when more than 2 alleles are found at a given locus38. Other genetic markers used include Cytochrome Oxidase subunit I sequence data44,45 or randomly amplified polymorphic DNA–PCR (RAPD–PCR) band patterns46. The detection of chimeric individuals may be improved with more markers, and whole-genome scanning techniques such as genotyping-by-sequencing (GBS) generate large amounts of genetic markers which can be applied to non model organisms47. In samples with scarce DNA material, a whole genome amplification (WGA) step is needed to obtain enough DNA48. The combination of WGA and GBS has been shown to reliably estimate multilocus genotypes in D. vexillum for clone detection and population genomics25 and can be an efficient and precise tool to assess chimerism.
In this study, we assess chimerism in D. vexillum and combine field surveys and experimental fusion tests with WGA-GBS genomic analyses from single zooids. Our objectives are (i) to report the prevalence of chimeric colonies in an introduced locality, (ii) describe the fusion/rejection behaviour between colony pairs, (iii) analyse the relation between colony fusion capability and genetic distance genomewide, and (iv) scan the dataset for candidate loci mediating colony fusion.
Methods
Two different approaches were followed to study the chimerism in Didemnum vexillum: (a) the identification of chimeric individuals in the wild, and (b) fusion experiments.
Sampling to identify chimeric individuals in the wild
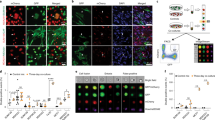
The first approach was carried out in oyster aquaculture facilities at the Fangar Bay (Ebro Delta, Spain, 40.776N, 0.737E). This system represents a favourable environment for D. vexillum in an enclosed area49,50. Nine colonies growing on commercial oysters were sampled. One central fragment and four peripheral fragments of 1 cm2, separated each other by at least 5 cm, were cut from each colony to determine the prevalence of chimeric colonies in this population (Fig. 1). The 45 fragments (5 for each of 9 colonies) were preserved in 96% ethanol for DNA extraction of a single zooid each.
Sampling scheme of a colony of D. vexillum growing over commercial oysters in the Ebro Delta for the prevalence study. Arrows indicate the 5 sampled fragments: one central fragment (1) and four peripheral fragments (2–5). Fragments 4 and 5 are next to fingers and its position is not visible due to the 3D structure of the colony.
Sampling and fusion experiments
In the second approach, a colony fusion experiment was carried out at the Venetian Lagoon with 3 sets of 5 colonies each (Fig. 2). This location is well suited for experimental work because it can be accessed directly from the laboratory facilities of the Institute of Marine Sciences (CNR-ISMAR). From each colony, identified with letters from K to Y, a fragment of <20 cm2 was cut into 7 pieces of 1–2 cm2 with a scalpel. One fragment was preserved in 96% ethanol for DNA extraction, two were paired with each other (intracolony pair), and the other 4 were paired to another colony fragment of the same set (intercolony pairs). The fragments used in this experiment were peripheral whenever possible, as it is described that they reattach faster than central fragments29. In total, there were 45 pairs, corresponding to 5 intracolony and 10 intercolony pairs for each set. All manipulation was done in the laboratory within hours of collection, and taking care to keep the colonies submerged in freshly collected lagoon water at all times.
Sampling map of the 15 colonies used in the fusion experiment carried out in the Venetian Lagoon. Each colour represents a different set. The scale bar represents 1 km. The asterisk identifies the spot in the Arsenale where the slides for the fusion experiment were submerged. The map was obtained and modified from Google Maps.
Each colony pair was fixed on a glass slide using cotton threads, with contacting cut edges to trigger a fast fusion/non-fusion outcome. The slides were submerged in vertical position at 2 meters depth at the ISMAR docks located in the ancient harbour of the Arsenal of Venice (Fig. 2). The study was carried out for 30 days, within the May-June period, coinciding with the growing season of D. vexillum in the region51. The slides were photographed twice a week to track colonies’ growth and fusion behaviour.
DNA extraction, sequencing, and loci identification
A thorax of a single zooid from each sample was dissected under the binocular and used for DNA extraction. We used an individual zooid to make sure we had a genetically homogeneous unit, and selected the thorax to avoid contaminating DNA from gut contents and to prevent a mixture of somatic and reproductive tissues. To get enough DNA from single thoraxes, a whole genome amplification (WGA) procedure was carried out with the REPLI-g® Single Cell kit (Qiagen) following manufacturer’s protocol except for the use of 1.5 µL of polymerase instead of the recommended 2 µL25. All samples were sent to the National Center of Genomic Analysis (CNAG, Barcelona) where a genotyping-by-sequencing (GBS) protocol was carried out. DNA was digested by PstI restriction enzyme and a paired-end sequencing of 2 × 125 bp fragments was performed in an Illumina HiSeq2500 platform.
Sequence quality filtering and loci identification and selection was carried out using the GIbPSs toolkit52 and following the same pipeline described in a previous study25. In short, the sequence quality filtering included the elimination of lower quality last bases by truncation and removal of reads with a low average Phred score threshold. The loci identification was divided in two steps: per sample and globally. First, sequences were analysed separately by sample, grouping identical reads into sequence variants and then into loci by pairwise comparisons. Second, a global locus and allele identification was performed to construct the loci dataset. The last main stage was the loci filtering where loci with alleles that could be indel variants, deeply sequenced loci and loci with more than two alleles per sample were removed. After these filters, loci shared by less than 70% of the samples were also deleted. The samples of Ebro Delta and Venice were analyzed separately to get two final loci datasets since they are significantly genetically differentiated populations25.
Data analysis
For each loci dataset, a table of haplotypic genotypes (i.e. alleles defined combining all variable positions of each locus) and a genepop file were exported from GIbPSs. The table of genotypes was used to get the Percentage of Shared Genotypes (PSG), defined as the percentage of identical genotypes among shared loci25. The PSG was calculated in R53, plotted using ‘ggplot2’54 and used to identify unique multilocus genotypes. Pairs of samples with PSG higher than 90% were considered the same genotype25. The genepop file was read into R using the ‘adegenet’ package55,56 and used to calculate the pairwise genetic Prevosti distance in ‘poppr’57,58 corrected by the exact number of loci shared by each pair.
The corrected Prevosti genetic distances between fused genotypes from the prevalence study and fused genotypes from the fusion experiment were compared with a Mann-Whitney-Wilcoxon test. For the fusion experiment, the corrected Prevosti distances between fused and non-fused intercolony pairs were compared with a Mann-Whitney-Wilcoxon test. The time to fuse between fused intercolony pairs and fused intracolony pairs was also compared with a Mann-Whitney-Wilcoxon test. A Pearson correlation coefficient between time to fuse and genetic distance was calculated for all fused pairs. All the analyses were run using R53.
For each locus of the fusion experiment dataset with less than 20% of missing data, the number of shared alleles (ranging from 0 to 2) of each surviving intercolony pair was calculated. Among them, we selected 1% of the loci with the highest absolute difference in shared alleles between fused and non-fused pairs. Consensus sequences of each selected locus were aligned using Blastn searches to the genomes of Ciona intestinalis (INSDC Assembly GCA_000224145.1; KH, Apr 2011) and C. savignyi (CSAV 2.0, Oct 2005) available on the Ensembl genome database59 (http://www.ensembl.org/index.html). Only hits with an E-value below 10–2 were considered.
Results
Prevalence study
An average of 2,510,418 raw reads per sample were obtained in the prevalence study at the Ebro Delta and 79,1% remained after the sequence quality filtering stage. A total of 69,600 loci were found among the 45 samples of which 2,145 polymorphic loci with 6,602 alleles were shared by at least 70% of the samples (Supplementary Dataset S1). The number of loci per sample averaged 1,995 but one sample shared only 536 loci (25% of the total loci), and was removed for further analyses.
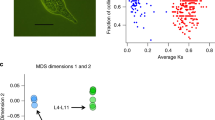
PSG clearly identified pairs of samples with the same genotype, which had identical alleles in 96.83 ± 0.14% (mean ± SE) of the loci, and pairs of samples with different genotypes (48.07 ± 0.07%) (Fig. 3). Based on these differences, we identified 13 unique genotypes among the 44 assayed zooids from the 9 colonies sampled at the Ebro Delta: 3 colonies showed 1 genotype, 3 colonies had 2 genotypes and 1 colony had 3 different genotypes. Additionally, 2 colonies presented the same genotype and were therefore identified as clones. Thus, 4 out of 9 sampled colonies were chimeric which implies that in the introduced population of the Ebro Delta we found 44% prevalence of chimerism. The mean corrected Prevosti genetic distance among genotypes in the chimeric colonies was 0.216 (SE ± 0.005).
Fusion experiment
We obtained an average of 2,127,134 raw reads per sample in the fusion experiment at the Venice lagoon and, after the quality filtering stage, 77.9% sequences were retained. We found a total of 44,688 loci of which 2,597 were polymorphic and shared by at least 70% of the samples (Supplementary Dataset S2). The total number of alleles was 7,343 and the mean number of loci per sample was 2,362. PSG values between colonies averaged 46.29% (SE ± 0.24), thus each sample had a distinct genotype and no clones were detected.
After the experiment set up, although the fragments were placed with contacting cut surfaces, most of them had to reattach, seal the cut, and grow into contact again. Some fragments reattached completely, while others only reattached partially, producing dead sections (Fig. 4). These dead sections were carefully removed as soon as they were detected. However, from the total of 45 pairs (15 intra- and 30 intercolony pairs), one or both fragments from 20 pairs (7 intra- and 13 intercolony pairs) could not reattach to the slide and thus died before any contact. No specific fragment typology (i.e. growing edge or central fragment) nor other external characteristics (i.e. colouring, thickness, roughness) seemed to correlate with mortality. All 8 surviving intracolony pairs and 5 intercolony pairs fused, while 11 intercolony pairs did not fuse. The number of fused, non-fused and dead pairs between the three different experimental sets were not significantly different (chi-squared = 2.65; p = 0.85).
Progression of 3 representative pairs of colonies during the fusion experiment showing the different behaviours observed. Pair number 25 (two fragments of colony R) corresponds to a fused intracolony pair, and numbers 5 (colonies K and O) and 27 (colonies R and T) are non-fused intercolony pairs. Black arrows indicate dead sections (before and after removal). White arrows show a complete fusion. Red arrows point out rejection with fragment retreat outcome. Blue arrows indicate non-fusion front formation. All photos are scaled and cropped to fit approximately the supporting slides, which are 25 × 75 mm. Photographic sequences are selected to illustrate the variety of possible outcomes, however, the experiments continued and lasted for 30 days.
We observed two different fusion phenotypes: complete fusion and partial fusion with some non-fusion front formation. All 8 surviving intracolony pairs fused completely (Fig. 4) and the limit between each fragment was rapidly blurred. On the contrary, only one out of the 5 intercolony pairs that fused did it completely and rapidly, while the others showed different behaviours. Two pairs fused only partially and formed a non-fusion front in a portion of the contacted edges. Another pair contacted at two separated points, in one of them colonies fused and in the other one of the colonies retreated. The last fused intercolony pair had a more complex behaviour, after one week of reattachment and growth, colonies met and fused at one contact point. However, a week later one of the colonies regressed partially at this point, while contact occurred again at a second point, where no fusion occurred. Finally, both colonies fused again at a third point of contact two weeks later.
We also observed two different non-fusion phenotypes: rejection with stable non-fusion front formation, and rejection with regression. All 11 non-fused colonies were intercolony pairs. Most of them (7), presented a non-fusion front all along the contacted region (Fig. 4). The other 4 pairs also formed an initial non-fusion front, only incipient in two of them, but one of the colonies finally retreated from the contact margin (Fig. 4).
The comparison between the mean corrected Prevosti distances of fused intercolony pairs (0.231 ± 0.002) and non-fused intercolony pairs (0.239 ± 0.003) showed no significant difference (Mann-Whitney-Wilcoxon = 42; p = 0.110) (Fig. 5). The time from first day of contact to first day of fusion between intracolony (0 to 4 days) and intercolony pairs (0 to 6 days) was not significantly different either (Mann-Whitney-Wilcoxon = 29; p = 0.15). Likewise, the correlation between time (days) to fuse and Prevosti genetic distances was not significant (Pearson correlation = 0.44; p = 0.13). On the other hand, the difference in the mean corrected Prevosti genetic distances among fused pairs (0.231 ± 0.002) and among genotypes fused to form the detected chimeras in the prevalence study (0.216 ± 0.005) was marginally significant (Mann-Whitney-Wilcoxon = 4; p = 0.052).
Candidate loci mediating colony fusion
From the total 2,597 loci of the fusion experiment dataset, 1,456 showed less than 20% of missing data. A total of 15 loci (1%) were considered as possible candidate loci for mediating colony fusion (Fig. 6; Supplementary Dataset S3) based on having the highest absolute difference between the mean number of shared alleles of fused pairs and non-fused pairs. In all 15 loci, this difference was greater than 0.85 and positive indicating that more shared alleles were found in fused pairs.
Absolute difference between mean number of shared alleles per locus among fused and non-fused intercolony pairs. Only loci with less than 20% of missing data are represented (N = 1,456). Solid dots correspond to the 1% loci with highest difference (>0.85). Loci are ordered by ID number in the x axis.
Among the 15 Blastn searches performed against the Ciona intestinalis genome, we found 3 hits although with low query cover and identity values. One was aligned to an intron of a predicted gene corresponding to an uncharacterized protein from chromosome 2. Another was aligned to an exon of a predicted gene coding for an uncharacterized protein from chromosome 8. The third locus was aligned to an intron of the gonadotropin releasing hormone receptor 1 (gnrhr1) gene from chromosome 3 (Table 1). Among the 15 Blastn searches against C. savignyi genome, we found only 3 hits, with similar query cover and identity values as for C. intestinalis. Only one locus aligned to a gene region corresponding to an intron of a predicted gene (Table 1).
Discussion
In colonial species, natural chimerism implies the presence of zooids with different genotypes within the same colony. We found a prevalence of 44% of chimeras in the Didemnum vexillum population of the Ebro Delta and 31% of intercolony pairs fused in the Venice experiment. The mean genetic distance between chimera-forming genotypes and experimentally fused colonies was 0.22 and 0.23 respectively. Both localities are heavily invaded by D. vexillum50,51, and the populations are genetically diverse and well differentiated25. In other species of tunicates the outcome of contacts between colonies is genetically regulated60. However, in our experiments, the genetic distances between fused genotypes were not significantly different than between non-fused genotypes, and we could not ascribe a clear role to the possible candidate loci detected. Therefore, in our genome-wide scan we could not identify the relevant loci for the allorecognition reaction between different colonies, if any.
Most of the genetic methods used for chimera detection underestimate the actual prevalence of chimerism due to the polymorphism of the markers used13. Next-generation sequencing (NGS) technologies such as GBS47 provide a tool to obtain large number of loci from species lacking a reference genome, producing reliable data to identify chimeras. In samples with scarce genetic material or species with reduced individual size, the combination of a WGA method with GBS proved robust and effective25 to obtain a large panel of loci that can be used to assess chimerism. In previous studies, the Percentage of Shared Genotypes (PSG) has been used to identify clones25 and in the present study it proved to be a fast and easy tool to detect chimeras. When zooids of the same genotype were compared, ca. 97% of the loci had the same genotypes and the 3% discrepancies can be explained by amplification and sequencing artifacts which could not be filtered during the bioinformatic analysis61. PSG dropped to less than 50% when colonies with different genotypes were compared. Note here that the relevant point is the existence of such a marked gap in the percentage of shared genotypes, as the precise mean PSG values may differ between studies depending on technical aspects (e.g., the number of markers, the stringency of the filters, or the artifacts during the process)61.
Natural chimerism has been found in several compound ascidians, but its prevalence varies considerably between and within species, from 0.5 to 39% in Botryllus schlosseri38,39,40,42, 1.9% in Botrylloides nigrum45, 1% in Perophora japonica44, from 3 to 61% in Diplosoma listerianum23, and from 17 to 48% in New Zealand populations of D. vexillum62. Our 44% prevalence of chimerism in the invasive population of the Ebro Delta is in the upper range of values found in other species, but low within those found in introduced populations of D. vexillum. In cut surface fusion experiments, an 80% of chimerism has been reported using colonies from the introduced area in New Zealand against only 27% in the native range32. Our experimental results in the Venetian Lagoon (31% fusion) are closer to the values found in the native range and far from those reported in New Zealand. This discrepancy may be explained by the different origin and genetic composition of the introduced populations in Europe and New Zealand25. More populations of this species should be studied to assess the extent of chimerism associated to introduced populations.
In species where a vascular system allowing an exchange of cells within colonies exists, a well-developed allorecognition system may reduce fitness costs of chimerism associated with somatic and germ-cell parasitism17,18,46,63. Didemnid ascidians such as D. listerianum and D. vexillum lack a colony-wide vascular system and zooids are only connected by the tunic, greatly decreasing the exchange of cells and hence the costs of chimerism. This type of colonial species may have a reduced or even absent allorecognition system, favouring more indiscriminate fusion between different colonies and making chimerism a more common condition46. Similarly, the relatedness of fused colonies also varies among species. In Botryllus spp. fusion can occur only between kin colonies39, while in D. listerianum46 and D.vexillum (present study) no genetic control mechanism has been detected and fusion takes place between non-related colonies. However, a study on D. vexillum described accumulation of diverse cell types in the tunic adjacent to allogeneic fusion areas, mostly phagocytes and morula cells64. Thus, it is likely that these cell types mediate the recognition reaction in this species, and that some limited exchange of tunic cells occurs between interacting colonies.
The most common outcomes described in fusion experiments among fragments of colonial species are fusion or rejection. However, more complex patterns have also been reported in most species. For instance, in the hydrozoan Hydractinia symbiolongicarpus four types of allorecognition phenotypes can be observed: fusion, rejection and two types of transitory fusion11. The stony coral Stylophora pistillata shows eight types of allorecognition reactions between kin colonies65. In the ascidians Botryllus schlosseri and Diplosoma listerianum, many different possible outcomes have been described22,41,66. In colonies of the ascidian Trididemnum solidum observed in natural environments, fusion has been observed after weeks or months of non-fused contacted margins from different colonies34. In a previous study on D. vexillum, highly dynamic interactions were found when allogeneic colonies came into contact33 and in many cases, fusion was followed by active growth away from fusion zones. The fragments resulting from this retreat showed predominantly segregated genotypes and chimeras were therefore transient in a scale of a few (10–12) days. In the present study, D. vexillum showed four different allorecognition phenotypes: complete fusion, partial fusion with some non-fusion front formation, rejection with stable non-fusion front formation, and rejection with regression. Some interactions were highly dynamic showing a combination of outcomes as the colonies contacted multiple times with a different outcome each time. However, fused colonies remained so until the end of the observation (30 days). Similarly, the finding of large chimeric colonies in the field (Ebro Delta) indicated that chimeras were not just transient, but stable entities. The external appearance of the colonies of D. vexillum and their behaviour in fusion experiments are different when comparing the photographic material from the present and previous studies29,33. We kept our experimental colonies in the natural environment of the lagoon while these previous studies were made under laboratory conditions. In the present work, colonies looked thicker and less transparent, suggesting that fragments may grow healthier under natural conditions.
Among our large loci dataset, none of the loci that we considered as possible candidates to mediate fusibility blasted to known genes with functions relevant for the allorecognition mechanisms. GBS produces huge numbers of loci randomly distributed through the genome of the target species47. Depending on the enzyme and species used, the studied loci may be differentially distributed among coding and non-coding regions67. For D. vexillum and using the PstI restriction enzyme, only 20% of the searched loci had a blast hit in Ciona genomes. Moreover, the query covers were small (~30%) and the E-value large suggesting that most of the analysed loci in this species will be located in non-coding regions and thus hard to identify in distant genomes. Thus, although we could not apparently detect any locus either directly involved in histocompatibility or linked to the relevant ones that could reveal a genetic control of the fusion/non fusion mechanism, we cannot discard that some of the identified loci are associated to highly specific regions mediating allorecognition. Moreover, the contrasting allorecognition phenotypes in fused isogeneic and allogeneic colonies (i.e., complete fusion in the former and complex, partial fusion patterns in the latter) might be indicative of a histocompatibility mechanism mediating fusibility in this species.
D. vexillum shows multiple advantageous biological traits that make it an aggressive invasive species that has colonized temperate regions worldwide. It has a rapid growth rate, produces large numbers of short-lived planktonic larvae and lacks significant predators (but see68,69). The importance of chimerism in adaptation and invasiveness can differ between species13. In the non-invasive but worldwide distributed populations of the branching coral Pocillopora damicornis, high levels of chimerism were found in extremely variable and highly impacted habitats70, suggesting chimerism may be involved in the success of adaptation of the species. In corals, chimerism has been suggested as an evolutionary mechanism of resilience and adaptation to global climate change impacts6. Similarly, chimerism has also been proposed as a driving factor in invasion success13,44. In colonial species with high growth-shrinkage dynamism and high fusion rates, chimerism may be a key aspect of the fragment dynamics and needs to be assessed for an efficient management62. Moreover, in the ascidian B. schlosseri, invasive populations seem to show higher levels of chimerism than in their native range13, and the same has been suggested for D. vexillum32. Chimeric colonies may show the ability to shift to advantageous genotypes in changing environments4. The invasive populations of D. vexillum assessed in the present study, the Ebro Delta and the Venetian Lagoon, show high levels of prevalence of chimerism and fusion rates, respectively. Thus, the role of chimerism in the success of the worldwide expansion of D. vexillum cannot be underestimated.
References
Buss, L. W. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. USA 79, 5337–5341 (1982).
Brusini, J., Robin, C. & Franc, A. To fuse or not to fuse? An evolutionary view of self-recognition systems. J. Phylogenetics Evol. Biol. 1, 1–8 (2013).
Pineda-Krch, M. & Lehtilä, K. Costs and benefits of genetic heterogeneity within organisms. J. Evol. Biol. 17, 1167–1177 (2004).
Rinkevich, B. Quo vadis chimerism? Chimerism 2, 1–5 (2011).
Barki, Y., Gateño, D., Graur, D. & Rinkevich, B. Soft-coral natural chimerism: a window in ontogeny allows the creation of entities comprised of incongruous parts. Mar. Ecol. Prog. Ser. 231, 91–99 (2002).
Rinkevich, B. Coral chimerism as an evolutionary rescue mechanism to mitigate global climate change impacts. Glob. Chang. Biol. 25, 1198–1206 (2019).
De Tomaso, A. W. et al. Isolation and characterization of a protochordate histocompatibility locus. Nature 438, 454–459 (2005).
Fernàndez-Busquets, X. & Burger, M. M. Cell adhesion and histocompatibility in sponges. Microsc. Res. Tech. 44, 204–218 (1999).
Blanquer, A. & Uriz, M. J. “Living together apart”’: the hidden genetic diversity of sponge populations. Mol. Biol. Evol. 28, 2435–2438 (2011).
Hughes, R. N., Manriquez, P. H., Morley, S., Craig, S. F. & Bishop, J. D. D. Kin or self-recognition? Colonial fusibility of the bryozoan Celleporella hyalina. Evol. Dev. 6, 431–437 (2004).
Powell, A. E. et al. Differential effect of allorecognition loci on phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa). Genetics 177, 2101–2107 (2007).
Rinkevich, B., Porat, R. & Goren, M. Allorecognition elements on a urochordate histocompatibility locus indicate unprecedented extensive polymorphism. Proc. R. Soc. London B 259, 319–324 (1995).
Ben-Shlomo, R. Invasiveness, chimerism and genetic diversity. Mol. Ecol. 26, 6502–6509 (2017).
Stoner, D. S., Rinkevich, B. & Weissman, I. L. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc. Natl. Acad. Sci. USA 96, 9148–9153 (1999).
Rinkevich, B. Natural chimerism in colonial urochordates. J. Exp. Mar. Bio. Ecol. 322, 93–109 (2005).
Scofield, V. L., Schlumpberger, J. M. & Weissman, I. L. Colony specificity in the colonial tunicate Botryllus and the origins of vertebrate immunity. Am. Zool. 22, 783–794 (1982).
Pancer, Z., Gershon, H. & Rinkevich, B. Coexistence and possible parasitism of somatic and germ cell lines in chimeras of the colonial urochordate Botryllus schlosseri. Biol. Bull. 189, 106–112 (1995).
Magor, B. G., De Tomaso, A. W., Rinkevich, B. & Weissman, I. L. Allorecognition in colonial tunicates: protection against predatory cell lineages? Immunol. Rev. 167, 69–79 (1999).
Rinkevich, B. & Yankelevich, I. Environmental split between germ cell parasitism and somatic cell synergism in chimeras of a colonial urochordate. J. Exp. Biol. 207, 3531–3536 (2004).
Kürn, U., Rendulic, S., Tiozzo, S. & Lauzon, R. J. Asexual propagation and regeneration in colonial ascidians. Biol. Bull. 221, 43–61 (2011).
Rinkevich, B. & Weissman, I. L. Chimeras vs genetically homogeneous individuals: potential fitness costs and benefits. Oikos 63, 119–124 (1992).
Bishop, J. D. D. & Sommerfeldt, A. D. Not like Botryllus: indiscriminate post-metamorphic fusion in a compound ascidian. Proc. R. Soc. London B 266, 241–248 (1999).
Sommerfeldt, A. D., Bishop, J. D. D. & Wood, C. A. Chimerism following fusion in a clonal ascidian (Urochordata). Biol. J. Linn. Soc. 79, 183–192 (2003).
Roman, J. & Darling, J. A. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464 (2007).
Casso, M., Turon, X. & Pascual, M. Single zooids, multiple loci: independent colonisations revealed by population genomics of a global invader. Biol. Inv, https://doi.org/10.1007/s10530-019-02069-8 (2019).
Bullard, S. G. et al. The colonial ascidian Didemnum sp. A: Current distribution, basic biology and potential threat to marine communities of the northeast and west coasts of North America. J. Exp. Mar. Bio. Ecol. 342, 99–108 (2007).
Mercer, J. M., Whitlatch, R. B. & Osman, R. W. Potential effects of the invasive colonial ascidian (Didemnum vexillum Kott, 2002) on pebble-cobble bottom habitats in Long Island Sound, USA. Aquat. Invasions 4, 133–142 (2009).
Lacoste, E. & Gaertner-Mazouni, N. Biofouling impact on production and ecosystem functioning: a review for bivalve aquaculture. Rev. Aquac. 7, 187–196 (2015).
Rinkevich, B. & Fidler, A. E. Initiating laboratory culturing of the invasive ascidian Didemnum vexillum. Manag. Biol. Invasions 5, 55–62 (2014).
Morris, J. A. Jr & Carman, M. R. Fragment reattachment, reproductive status, and health indicators of the invasive colonial tunicate Didemnum vexillum with implications for dispersal. Biol. Invasions 14, 2133–2140 (2012).
Reinhardt, J. F. et al. Material properties of Didemnum vexillum and prediction of tendril fragmentation. Mar. Biol. 159, 2875–2884 (2012).
Smith, K. F. et al. Increased inter-colony fusion rates are associated with reduced COI haplotype diversity in an invasive colonial ascidian Didemnum vexillum. PLoS One 7, e30473 (2012).
Fidler, A. E., Bacq-Labreuil, A., Rachmilovitz, E. & Rinkevich, B. Efficient dispersal and substrate acquisition traits in a marine invasive species via transient chimerism and colony mobility. PeerJ 6, e5006 (2018).
Bak, R. P. M., Sybesma, J. & Duyl, F. C. Van. The ecology of the tropical compound ascidian Trididemnum solidum. II. Abundance, Growth and Survival. Mar. Ecol. Prog. Ser. 6, 43–52 (1981).
Westerman, E. L., Dijkstra, J. A. & Harris, L. G. High natural fusion rates in a botryllid ascidian. Mar. Biol. 156, 2613–2619 (2009).
López-Legentil, S., Erwin, P. M., Velasco, M. & Turon, X. Growing or reproducing in a temperate sea: optimization of resource allocation in a colonial ascidian. Invertebr. Biol. 132, 69–80 (2013).
Watanabe, H. & Taneda, Y. Self or non-self recognition in compound ascidians. Am. Zool. 22, 775–782 (1982).
Ben-Shlomo, R., Douek, J. & Rinkevich, B. Heterozygote deficiency and chimerism in remote populations of a colonial ascidian from New Zealand. Mar. Ecol. Prog. Ser. 209, 109–117 (2001).
Ben-Shlomo, R., Motro, U., Paz, G. & Rinkevich, B. Pattern of settlement and natural chimerism in the colonial urochordate Botryllus schlosseri. Genetica 132, 51–58 (2008).
Ben-Shlomo, R., Reem, E., Douek, J. & Rinkevich, B. Population genetics of the invasive ascidian Botryllus schlosseri from South American coasts. Mar. Ecol. Prog. Ser. 412, 85–92 (2010).
Stoner, D. S. & Weissman, I. L. Somatic and germ cell parasitism in a colonial ascidian: Possible role for a highly polymorphic allorecognition system. Proc. Natl. Acad. Sci. USA 93, 15254–15259 (1996).
Paz, G., Douek, J., Mo, C., Goren, M. & Rinkevich, B. Genetic structure of Botryllus schlosseri (Tunicata) populations from the Mediterranean coast of Israel. Mar. Ecol. Prog. Ser. 250, 153–162 (2003).
Schweinsberg, M., Weiss, L. C., Striewski, S., Tollrian, R. & Lampert, K. P. More than one genotype: how common is intracolonial genetic variability in scleractinian corals? Mol. Ecol. 24, 2673–2685 (2015).
Pérez-Portela, R., Turon, X. & Bishop, J. D. D. Bottlenecks and loss of genetic diversity: Spatio-temporal patterns of genetic structure in an ascidian recently introduced in Europe. Mar. Ecol. Prog. Ser. 451, 93–105 (2012).
Sheets, E. A., Cohen, C. S., Ruiz, G. M. & da Rocha, R. M. Investigating the widespread introduction of a tropical marine fouling species. Ecol. Evol. 6, 2453–2471 (2016).
Sommerfeldt, A. D. & Bishop, J. D. D. Random amplified polymorphic DNA (RAPD) analysis reveals extensive natural chimerism in a marine protochordate. Mol. Ecol. 8, 885–890 (1999).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379 (2011).
Dean, F. B. et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. 99, 5261–5266 (2002).
Casso, M. et al. Seasonal patterns of settlement and growth of introduced and native ascidians in bivalve cultures in the Ebro Delta (NE Iberian Peninsula). Reg. Stud. Mar. Sci. 23, 12–22 (2018).
Ordóñez, V. et al. Ongoing expansion of the worldwide invader Didemnum vexillum (Ascidiacea) in the Mediterranean Sea: high plasticity of its biological cycle promotes establishment in warm waters. Biol. Invasions 17, 2075–2085 (2015).
Tagliapietra, D., Keppel, E., Sigovini, M. & Lambert, G. First record of the colonial ascidian Didemnum vexillum Kott, 2002 in the Mediterranean: Lagoon of Venice (Italy). BioInvasions Rec. 1, 247–254 (2012).
Hapke, A. & Thiele, D. GIbPSs: a toolkit for fast and accurate analyses of genotyping-by-sequencing data without a reference genome. Mol. Ecol. Resour. 16, 979–990 (2016).
R Core Team. A Language and Environment for Statistical Computing. (2018).
Wickham, H. ggplot2: Elegant graphics for data analysis. (Springer-Verlag, 2009).
Jombart, T. adegenet: an R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Jombart, T. & Ahmed, I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Kamvar, Z. N., Brooks, J. C. & Grünwald, N. J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 6, 208 (2015).
Kamvar, Z. N., Tabima, J. F. & Grünwald, N. J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014).
Zerbino, D. R. et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018).
Weissman, I. L., Saito, Y. & Rinkevich, B. Allorecognition histocompatibility in a protochordate species: is the relationship to MHC somatic or structural? Immunol. Rev. 113, 227–241 (1990).
O’Leary, S. J., Puritz, J. B., Willis, S. C., Hollenbeck, C. M. & Portnoy, D. S. These aren’t the loci you’e looking for: Principles of effective SNP filtering for molecular ecologists. Mol. Ecol. 27, 3193–3206 (2018).
Watts, A. M., Hopkins, G. A. & Goldstien, S. J. Chimerism and population dieback alter genetic inference related to invasion pathways and connectivity of biofouling populations on artificial substrata. Ecol. Evol. 9, 3089–3104 (2019).
Paz, G. & Rinkevich, B. Morphological consequences for multi-partner chimerism in Botrylloides, a colonial urochordate. Dev. Comp. Immunol. 26, 615–622 (2002).
Sellers, A. E., Fagerberg, W. R. & Litvaitis, M. K. Tunic cell populations during fusion events in the colonial ascidian Didemnum vexillum (Tunicata). Invertebr. Biol. 132, 394–403 (2013).
Amar, K. O., Chadwick, N. E. & Rinkevich, B. Coral kin aggregations exhibit mixed allogeneic reactions and enhanced fitness during early ontogeny. BMC Evol. Biol. 8, 126 (2008).
Rinkevich, B. & Weissman, I. L. A long-term study on fused subclones in the ascidian Botryllus schlosseri: the resorption phenomenon (Protochordata: Tunicata). J. Zool. 213, 717–733 (1987).
Carreras, C. et al. Population genomics of an endemic Mediterranean fish: differentiation by fine scale dispersal and adaptation. Sci. Rep. 7, 43417 (2017).
Forrest, B. M., Fletcher, L. M., Atalah, J., Piola, R. F. & Hopkins, G. A. Predation limits spread of Didemnum vexillum into natural habitats from refuges on anthropogenic structures. PLoS One 8, 1–12 (2013).
Stefaniak, L. M. Mechanisms for invasion success by Didemnum vexillum (Chordata: Ascidiacea): observations versus manipulations. Biol. Invasions 19, 1213–1225 (2017).
Rinkevich, B., Shaish, L., Douek, J. & Ben-Shlomo, R. Venturing in coral larval chimerism: a compact functional domain with fostered genotypic diversity. Sci. Rep. 6, 19493 (2016).
Acknowledgements
We are grateful to Margarita Fernandez (IRTA) and Víctor Ordóñez for logistic help in the Ebro Delta. Carles Bori from granting access to his oyster culture facility. Marco Sigovini and Irene Guarneri (ISMAR/CNR) provided invaluable help during the experiments in Venice. Funding was obtained from project PopCOmics (CTM2017-88080, MCIU, AEI/FEDER/UE). This is a contribution from the Consolidated Research Group “Benthic Biology and Ecology” SGR2017-1120 (Catalan Government).
Author information
Authors and Affiliations
Contributions
M.C., M.P. and X.T. conceived and planned the study. M.C., M.P. and X.T. performed fieldwork at the Ebro Delta. M.C. and D.T. planned and executed the experiments in Venice. M.C. carried out the molecular lab and bioinformatics procedures. M.C., M.P. and X.T. conducted statistical analyses. M.C. drafted the initial manuscript. All authors revised the manuscript and contributed to data interpretation. All authors approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Casso, M., Tagliapietra, D., Turon, X. et al. High fusibility and chimera prevalence in an invasive colonial ascidian. Sci Rep 9, 15673 (2019). https://doi.org/10.1038/s41598-019-51950-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51950-y
This article is cited by
-
Prokaryotic symbiont communities in three ascidian species introduced in both Ireland and New Zealand
Environmental Science and Pollution Research (2023)
-
Genetic diversity and relatedness in aquaculture and marina populations of the invasive tunicate Didemnum vexillum in the British Isles
Biological Invasions (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.